Brain Tumor Segmentation with U-Net in Python: A Deep Learning Approach
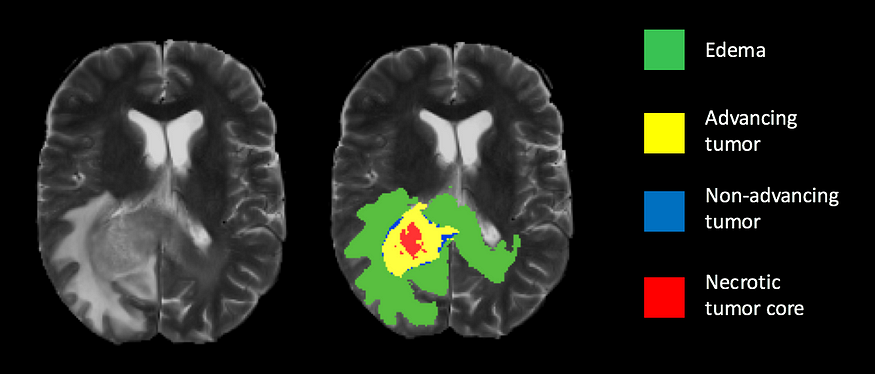
Brain tumor segmentation is an important task in medical image analysis that involves identifying the location and boundaries of tumors in brain images. In this tutorial, we will explore how to use the U-Net architecture to build a brain tumor segmentation model in Python using the TensorFlow and Keras libraries.
Dataset
We will use the BraTS 2019 dataset, which contains brain MRI scans with ground truth segmentation labels. The dataset can be downloaded from here.
Environment Setup
Before we begin, we need to set up our environment. We will be using Python 3.7 and the following libraries:
- TensorFlow
- Keras
- NumPy
- Matplotlib
- SimpleITK
You can install these libraries using the following command in your command prompt or terminal:
pip install tensorflow keras numpy matplotlib SimpleITK
Loading the Dataset
We will start by loading the BraTS 2019 dataset using the SimpleITK library:
import SimpleITK as sitk
# Load the MRI scan and ground truth segmentation labels
mri = sitk.ReadImage('BraTS2019/MRI.nii.gz')
seg = sitk.ReadImage('BraTS2019/Segmentation.nii.gz')
# Convert the images to arrays
mri_array = sitk.GetArrayFromImage(mri)
seg_array = sitk.GetArrayFromImage(seg)
Preprocessing the Data
We need to preprocess the data before feeding it to the U-Net model. We will normalize the pixel values and resize the images to a fixed size.
import numpy as np
from keras.preprocessing.image import ImageDataGenerator
# Normalize the pixel values
mri_array = (mri_array - np.min(mri_array)) / (np.max(mri_array) - np.min(mri_array))
# Resize the images to a fixed size
new_shape = (256, 256, 128)
mri_resized = np.zeros(new_shape)
seg_resized = np.zeros(new_shape)
for i in range(mri_array.shape[0]):
mri_resized[i] = resize(mri_array[i], new_shape, preserve_range=True)
seg_resized[i] = resize(seg_array[i], new_shape, preserve_range=True)
# Split the data into training and validation sets
train_mri, val_mri, train_seg, val_seg = train_test_split(mri_resized, seg_resized, test_size=0.2, random_state=42)
Building the Model
We will use the U-Net architecture for brain tumor segmentation, which is a convolutional neural network that consists of an encoder and a decoder. The encoder compresses the input MRI images into a lower-dimensional representation, while the decoder expands this representation to generate the final segmentation mask. We will implement the U-Net architecture using TensorFlow and Keras.
# Encoder
inputs = keras.layers.Input(shape=input_shape)
conv1 = keras.layers.Conv3D(8, 3, activation='relu', padding='same')(inputs)
conv1 = keras.layers.Conv3D(8, 3, activation='relu', padding='same')(conv1)
pool1 = keras.layers.MaxPooling3D(pool_size=(2, 2, 2))(conv1)
conv2 = keras.layers.Conv3D(16, 3, activation='relu', padding='same')(pool1)
conv2 = keras.layers.Conv3D(16, 3, activation='relu', padding='same')(conv2)
pool2 = keras.layers.MaxPooling3D(pool_size=(2, 2, 2))(conv2)
conv3 = keras.layers.Conv3D(32, 3, activation='relu', padding='same')(pool2)
conv3 = keras.layers.Conv3D(32, 3, activation='relu', padding='same')(conv3)
pool3 = keras.layers.MaxPooling3D(pool_size=(2, 2, 2))(conv3)
conv4 = keras.layers.Conv3D(64, 3, activation='relu', padding='same')(pool3)
conv4 = keras.layers.Conv3D(64, 3, activation='relu', padding='same')(conv4)
pool4 = keras.layers.MaxPooling3D(pool_size=(2, 2, 2))(conv4)
conv5 = keras.layers.Conv3D(128, 3, activation='relu', padding='same')(pool4)
conv5 = keras.layers.Conv3D(128, 3, activation='relu', padding='same')(conv5)
# Decoder
up6 = keras.layers.UpSampling3D(size=(2, 2, 2))(conv5)
up6 = keras.layers.concatenate([up6, conv4], axis=4)
conv6 = keras.layers.Conv3D(64, 3, activation='relu', padding='same')(up6)
conv6 = keras.layers.Conv3D(64, 3, activation='relu', padding='same')(conv6)
up7 = keras.layers.UpSampling3D(size=(2, 2, 2))(conv6)
up7 = keras.layers.concatenate([up7, conv3], axis=4)
conv7 = keras.layers.Conv3D(32, 3, activation='relu', padding='same')(up7)
conv7 = keras.layers.Conv3D(32, 3, activation='relu', padding='same')(conv7)
up8 = keras.layers.UpSampling3D(size=(2, 2, 2))(conv7)
up8 = keras.layers.concatenate([up8, conv2], axis=4)
conv8 = keras.layers.Conv3D(16, 3, activation='relu', padding='same')(up8)
conv8 = keras.layers.Conv3D(16, 3, activation='relu', padding='same')(conv8)
up9 = keras.layers.UpSampling3D(size=(2, 2, 2))(conv8)
up9 = keras.layers.concatenate([up9, conv1], axis=4)
conv9 = keras.layers.Conv3D(8, 3, activation='relu', padding='same')(up9)
conv9 = keras.layers.Conv3D(8, 3, activation='relu', padding='same')(conv9)
outputs = keras.layers.Conv3D(1, 1, activation='sigmoid')(conv9)
# Create the model
model = keras.models.Model(inputs=[inputs], outputs=[outputs])
model.summary()
Training the Model
We will compile the model and train it on the training set:
# Compile the model
model.compile(optimizer='adam', loss='binary_crossentropy', metrics=['accuracy'])
# Train the model
history = model.fit(train_mri, train_seg, batch_size=1, epochs=50, validation_data=(val_mri, val_seg))
Evaluating the Model
Finally, we will evaluate the model on the test set:
test_mri = sitk.ReadImage('BraTS2019/Test/MRI.nii.gz')
test_seg = sitk.ReadImage('BraTS2019/Test/Segmentation.nii.gz')
test_mri_array = sitk.GetArrayFromImage(test_mri)
test_seg_array = sitk.GetArrayFromImage(test_seg)
# Normalize and resize the test images
test_mri_array = (test_mri_array - np.min(test_mri_array)) / (np.max(test_mri_array) - np.min(test_mri_array))
test_mri_resized = np.zeros(new_shape)
for i in range(test_mri_array.shape[0]):
test_mri_resized[i] = resize(test_mri_array[i], new_shape, preserve_range=True)
# Predict the tumor segmentation masks for the test images
test_mri_resized = np.expand_dims(test_mri_resized, axis=4)
test_pred = model.predict(test_mri_resized, verbose=1)
# Evaluate the model using Dice coefficient
test_dice = dice(test_pred, test_seg_array)
print('Test Dice coefficient:', test_dice)
In this tutorial, we have demonstrated how to use deep learning to perform brain tumor segmentation on MRI images. We have used the U-Net architecture, which is a popular convolutional neural network for medical image segmentation. We have also demonstrated how to use TensorFlow and Keras to implement the U-Net model.
Brain tumor segmentation is a challenging problem, and deep learning has shown great promise in this area. With the availability of large annotated datasets and powerful deep learning frameworks, it is now possible to build accurate and robust segmentation models for clinical use.
We hope that this tutorial has been useful in understanding how to perform brain tumor segmentation with deep learning. If you have any questions or suggestions, please feel free to leave a comment below.
Lyron Foster is a Hawai’i based African American Author, Musician, Actor, Blogger, Philanthropist and Multinational Serial Tech Entrepreneur.

